The X-chromosomal short tandem repeats analysis in Thai population
Wittawat Chantkran1 and Budsaba Rerkamnuaychoke2*
1 Department of Pathology, Phramongkutklao College of Medicine, Bangkok, Thailand
2 Human Genetics Unit, Department of Pathology, Faculty of Medicine Ramathibodi Hospital,
Mahidol University, Bangkok, Thailand
Correspondence to:
Associate Professor Dr Budsaba Rerkamnuaychoke,
Human Genetics Unit, Department of Pathology, Floor 4, Main Building,
Faculty of Medicine Ramathibodi Hospital, Mahidol University,
270 Rama VI Road, Rajadevi, Bangkok, 10400 Thailand.
Telephone: +66 (0) 2 201 1368 and +66 (0) 2 201 1369 Fax: +66 (0) 2 201 1464
Email: budsaba.rer @mahidol.ac.th, rabrk@mahidol.ac.th
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
Abstract
Human variations in the X chromosome are useful tools in studying human genetic diversity and individual identification. Five X chromosomal short tandem repeats (X-STRs) multiplex system (DXS8378, DXS101, HPRTB, DXS8377 and DXS10011) were amplified in one single polymerase chain reaction. DNA samples of 200 (101 males and 99 females) unrelated healthy individuals Thai, and 15 family trios with female children, were successfully analysed using this five X-STRs multiplex system. The distributions of allele frequencies were examined for independence. When the forensic efficiency was calculated, DXS10011 locus was found to be the greatest marker for forensic application and population study. The combined powers of discrimination of five loci in males and females were 0.999993 and 0.999999, respectively. These five X chromosome markers are highly informative for population study and the database of Thailand.
Keywords: X chromosome; short tandem repeats; multiplex PCR; Thai population; forensics
Introduction
For many years, autosomal and Y-chromosomal DNA have been used for forensic purposes(1,2). At the present time, X-chromosomal markers have been observed as a forensic of interest(3-7). The availability of the finished sequence of the human X chromosome(8), now allows exploring its evolution and unique properties at a new level. The X chromosome contains about 5% of the haploid genome and is completely conserved in gene content between species. However, evolutionary processes are likely to have shaped the behaviour and structure of the X chromosome in many other ways, influencing features such as repeat content, gene content, mutation rate and haplotype structure.
Study results of variations in the X chromosome are invaluable tools for studying the human genetic diversity and individual identification. The X-STRs may strengthen the results of autosomal and Y-STRs analysis. In paternity testing, although the X-STRs are advantageous only in case the alleged child is female, the X-STRs often have a greater power of exclusion than autosomal markers. Besides, the X-STRs are very beneficial in maternity testing or in deficiency paternity cases, for instance, to judge the paternity of disputed half-sisters with the same father in case the mothers' DNA are unavailable(5).
The X-chromosome is inherited via a sex-based pattern. Because the recombination of the X chromosome occurs only in females, there is a less effective population size, greater linkage disequilibrium, and a stronger genetic drift. These factors bring the X chromosome to be a source of data in human evolution and evacuation studies(8).
In this study, five X chromosome markers of interest in forensic science will be typed in the Thai population in order to report population database and haplotype profiling in the Thai population and calculating the population genetic parameters for these markers.
Materials and Methods
DNA extraction:
DNA extraction from whole blood samples of 200 Thai people (101 males and 99 females) were performed with a commercial method, according to manufacturer's instructions (Promega Corp., Madison, WI, USA). An aliquot containing approximately 4 ng of DNA was used in each PCR amplification. In addition, 15 family trios with female children (previously confirmed by autosomal STRs analysis) were checked for regular X-chromosomal inheritance.
STR amplification and fragment analysis:
After various tests with different STRs and optimisation of PCR parameters, the STRs DXS8378, DXS101, HPRTB, DXS8377 and DXS10011 were combined into the PCR multiplex system. Primer sequences, labeling, concentrations, and references are shown in Table 1. Besides the primers, the PCR reaction mix (12.5 µl reaction volume) contained 1X AmpliTaq buffer II, 1.5 mM of MgCl2, 200 µl of each dNTP, and 2 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster city, CA, USA). A GeneAmp PCR system 9700 thermal cycler (Applied Biosystems, Foster city, CA, USA) was used. After several pilot experiments to optimise the amplification of all markers in the same tube, a PCR touch-down protocol was chosen(9), consisting of an initial denaturation step at 95 oC for 10 min, followed by nine cycles with denaturation at 94 oC for 30 s, annealing at decreasing temperature between 61 and 65 oC (decreasing 0.5 oC each cycle) for 1 min 30 s and extension at 72 oC for 1 min 15 s. Then 28 cycles at 94 oC 30 s, 58 oC 1 min 30 s, 72 oC 1 min 15 s; followed by a final extension at 60 oC for 60 min.
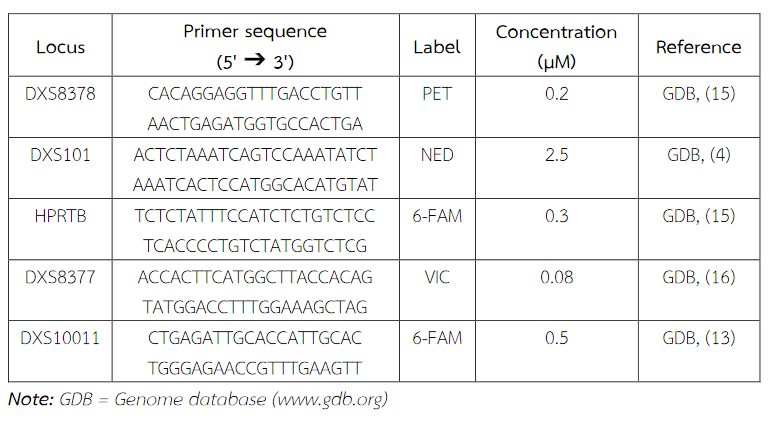
Table 1 Primer sequences, concentrations, dye labeling and references.
An aliquot containing 1 µl of PCR product was mixed with 10 µl of Hi-Di formamide and 0.5 µl of GeneScan 500LIZ size standard (Applied Biosystems), heated at 95 oC for 3 min, quenched at 4 oC for 3 min and injected into an Applied Biosystems 3130 Genetic Analyzer. Fragment sizes were automatically determined using GeneScan analysis software (Applied Biosystems). Genotyping was analysed using GeneMapper ID software (Applied Biosystems) by comparison with reference DNA control sample 9947A (female) (Applied Biosystems). Alleles were assigned according to the recommendations of the International Society of Forensic Haemogenetics (ISFH) commission(10).
Statistical analysis:
Allelic frequencies were calculated by using the data from males and females collectively and observed heterozygosity (HETobs) were calculated using the female data by Power Stat V12 program (www.promega.com). Mean exclusion chance (MEC), power of discrimination for females (PDf) and power of discrimination for males (PDm) were computed by chromosome X web software (www.chrx-str.org). The combined power of discrimination was determined as 1-[(1-PDa)•(1-PDb)•(1-PDc)•(1-PDd)•••] with PDa-d indicating the discriminating power of the different polymorphism(11). To evaluate Hardy-Weinberg Equilibrium (HWE) and linkage disequilibrium (LD), GENEPOP program (version 3.4) was used.
Results
The 9947A DNA (female) was used as a control reference sample. The allele frequencies for the X-chromosomal STRs DXS8378, DXS101, and HPRTB are displayed in Table 2, the data for DXS8377and DXS10011 in Table 3. The forensic efficiency of the five X-STRs loci were calculated (Table 4). DXS8378 had the lowest values for PDm, PDf, and MEC in the present study. The combined power of discrimination in males and females were 0.999993 and 0.999999, respectively. Observed heterozygosity (HETobs) in females for the five markers ranged from 0.660 to 0.900 in this study, DXS8377 had the highest whereas DXS8378 had the lowest HETobs.
An exact test for Hardy-Weinberg equilibrium (HWE) performed on female samples indicated that the genotype distributions did not deviate from HWE at any of the loci, except for threshold significance at locus DXS10011 (P=0.0000; data not shown). When exact tests showed no significant difference between allele distributions of the two groups, pooled male and female databases of the Thai populations were purposely created to compare the allele frequencies at locus DXS8378, DXS101, HPRTB, DXS8377, and DXS10011 between different races (Tables 5 – 9).
Six alleles were identified at locus DXS8378, ranging from allele 8 to 13, the most common being 10 (Table 2). Allele 10 was also observed as a most common allele shared among the Asian population (Thai, Japan and China) and allele 8 was found only in the Thai population (Table 5). Twelve alleles were found at locus DXS101, ranging from allele 18 to 30, the most common being 25 (Table 2). The comparison of allele frequencies at DXS101 locus between different races showed no significant most common allele shared in any population (Table 6). Five alleles ranging from allele 11 to 15 were identified at locus HPRTB, the most common allele being 13 (Table 2). Allele 13 was observed as a most common allele shared among the Asian population (Thai, Japan and China) (Table 7). DXS8377 had 17 alleles ranging from 41 to 57. The most common allele was 47 (Table 3). Among the African population (Angola, Uganda and Mozambique), a shared most common allele was 49 (Table 8). DXS10011 had 32 alleles ranging from 27 to 51, the most common allele was 38 (Table 3). There was no significant most common allele shared between different races at locus DXS10011 (Table 9).
Table 2 Allele frequencies for the X-chromosomal STRs DXS8378, DXS101 and HPRTB in Thai population. Allele frequencies were collected from the analysis of DNA samples of 200 (101 males and 99 females) unrelated healthy individuals Thai.
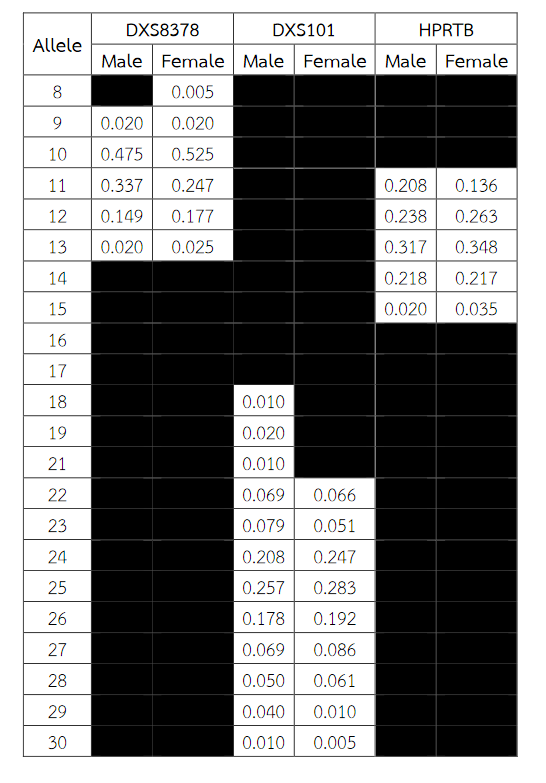
Table 3 Allele frequencies for the X-chromosomal STRs DXS8377 and DXS10011 in Thai population. Allele frequencies were collected from the analysis of DNA samples of 200 (101 males and 99 females) unrelated healthy individuals Thai.
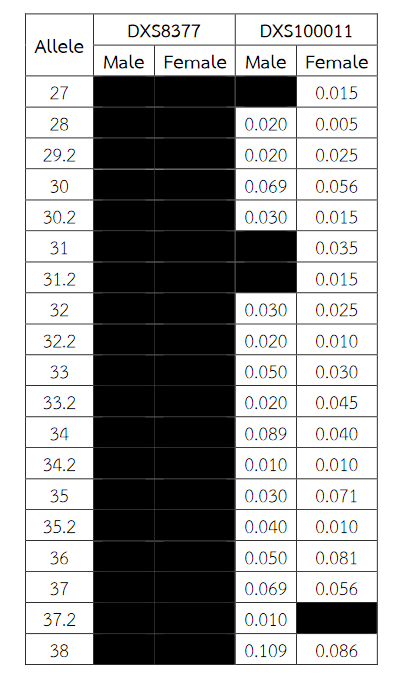
Table 3 (Continued) Allele frequencies for the X-chromosomal STRs DXS8377 and DXS10011 in Thai population. Allele frequencies were collected from the analysis of DNA samples of 200 (101 males and 99 females) unrelated healthy individuals Thai.
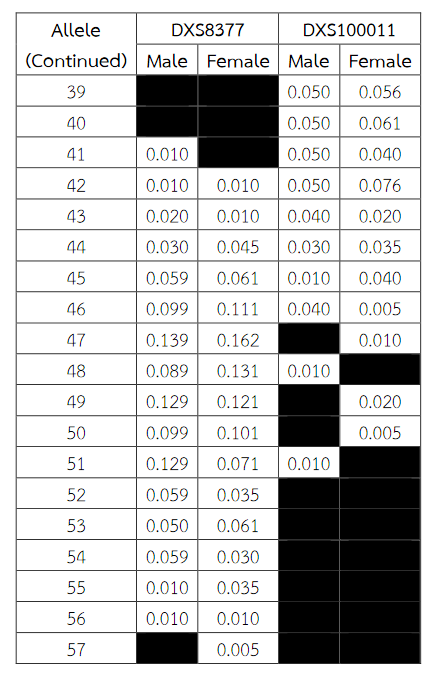
Table 4 Statistical parameters of forensic interest for X-chromosomal STRs in Thai population. Statistical parameters were analysed from 200 (101 males and 99 females) unrelated healthy individuals Thai shown in Table 2 and 3. HETobs, HETexp and PDf were analysed from females only. PDm was analysed from males only.
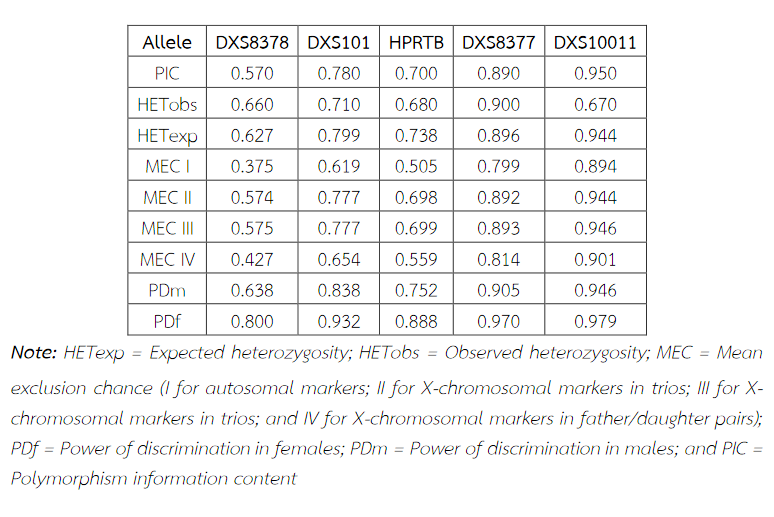
Table 5 Allele frequencies, statistical parameters and references of DXS8378 in six populations. For the Thai population, the data were from pooled males and females (Table 2) when the exact tests showed no difference between the two groups. Allele 10 was observed as a most common allele shared among the Asian population (Thai, Japan and China) and allele 8 was found only in Thai population. Some statistical parameters were not available.
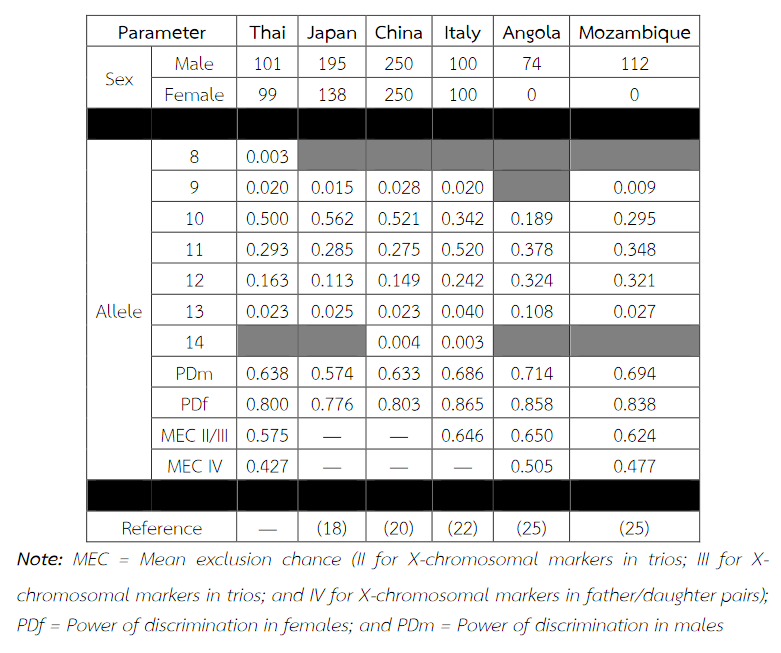
Table 6 Allele frequencies, statistical parameters and references of DXS101 in six populations. For the Thai population, the data were from pooled males and females (Table 2) when the exact tests showed no difference between the two groups. No significant most common allele shared in any population was observed. Some statistical parameters were not available.
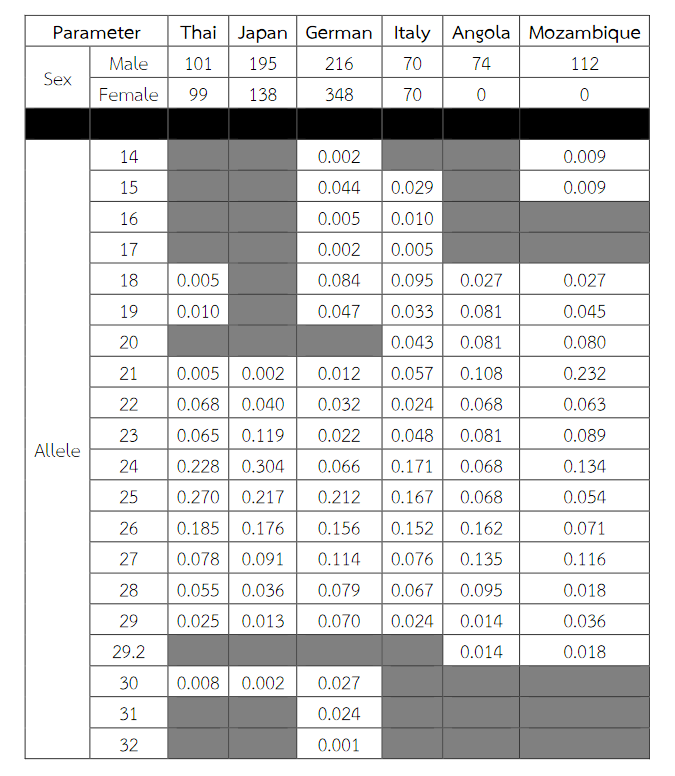
Table 6 (Continued) Allele frequencies, statistical parameters and references of DXS101 in six populations. For the Thai population, the data were from pooled males and females (Table 2) when the exact tests showed no difference between the two groups. No significant most common allele shared in any population was observed. Some statistical parameters were not available.
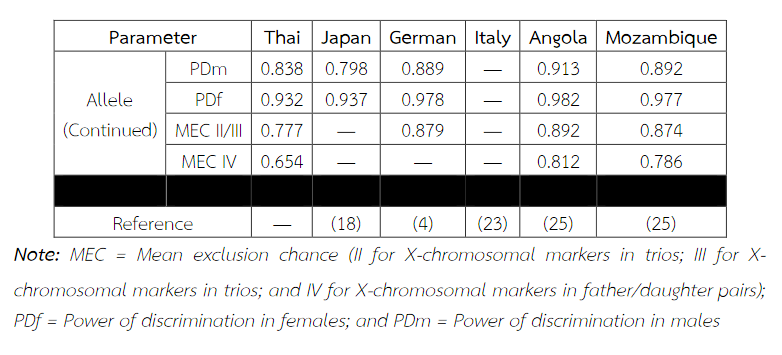
Table 7 Allele frequencies, statistical parameters and references of HPRTB in six populations. For the Thai population, the data were from pooled males and females (Table 2) when the exact tests showed no difference between the two groups. Allele 13 was observed as a most common allele shared among the Asian population (Thai, Japan and China). Some statistical parameters were not available.
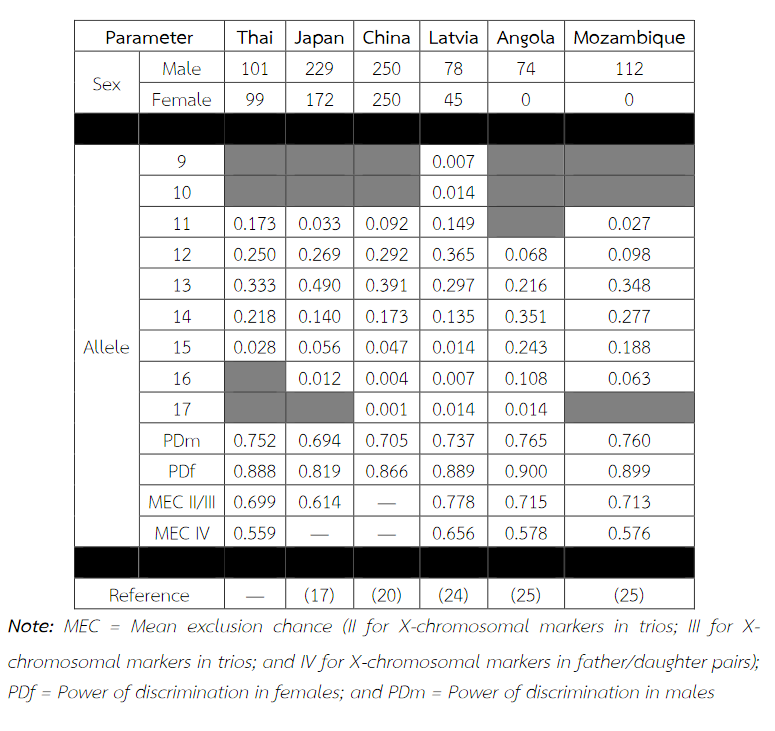
Table 8 Allele frequencies, statistical parameters, and references of DXS8377 in six populations. For the Thai population, the data were from pooled males and females (Table 3) when the exact tests showed no difference between the two groups. Allele 49 was observed as a most common allele shared among the African population (Angola, Uganda and Mozambique). Some statistical parameters were not available.
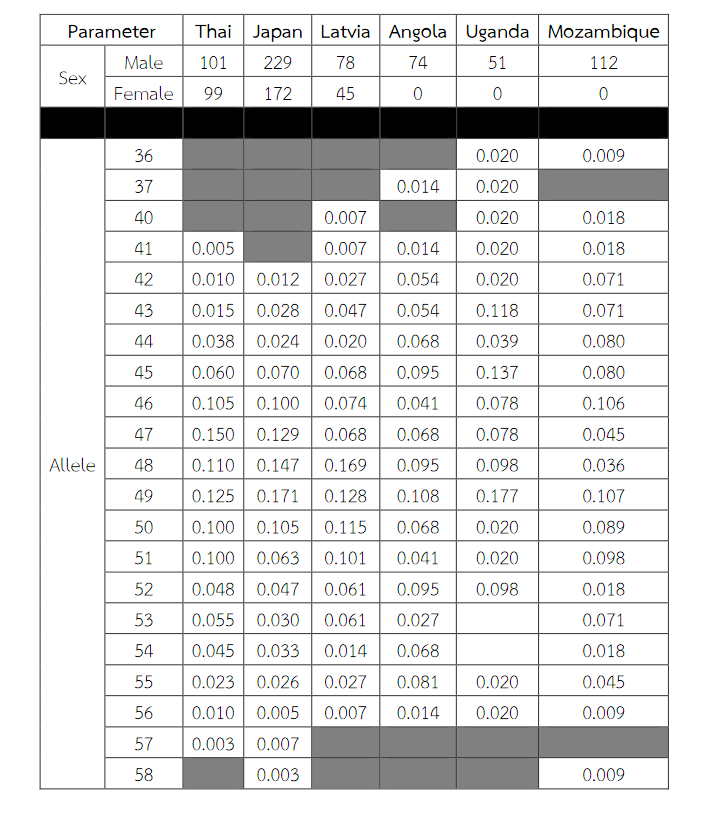
Table 8 (Continued) Allele frequencies, statistical parameters, and references of DXS8377 in six populations. For the Thai population, the data were from pooled males and females (Table 3) when the exact tests showed no difference between the two groups. Allele 49 was observed as a most common allele shared among the African population (Angola, Uganda and Mozambique). Some statistical parameters were not available.
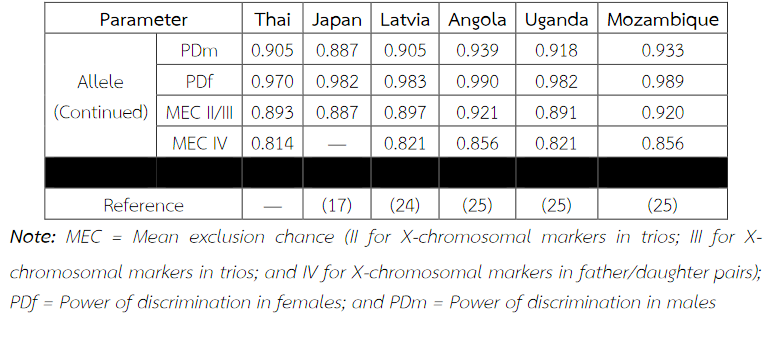
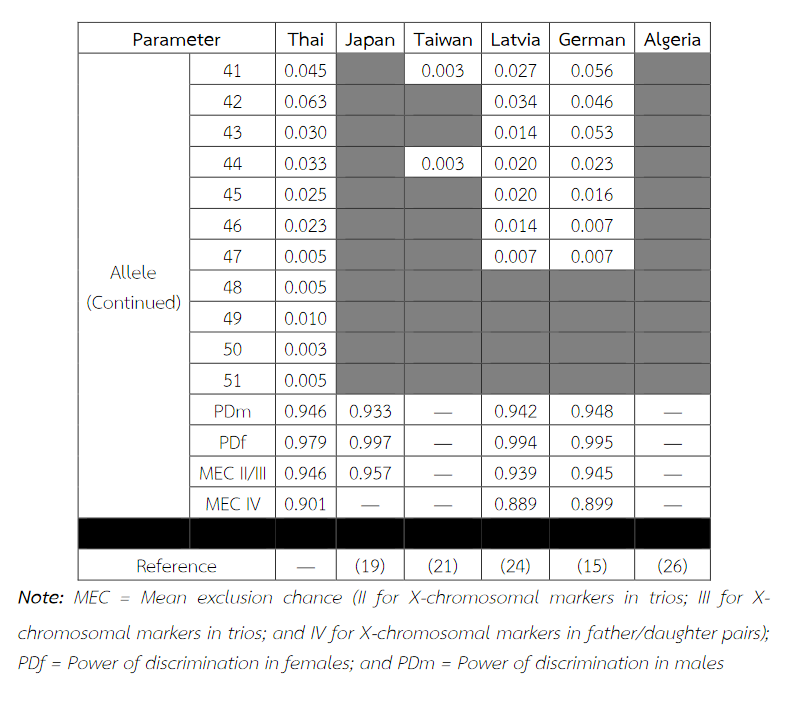
Table 9 Allele frequencies, statistical parameters, and references of DXS10011 in six populations. For the Thai population, the data were from pooled males and females (Table 3) when the exact tests showed no difference between the two groups. No significant most common allele shared in any population was observed. Some statistical parameters were not available.
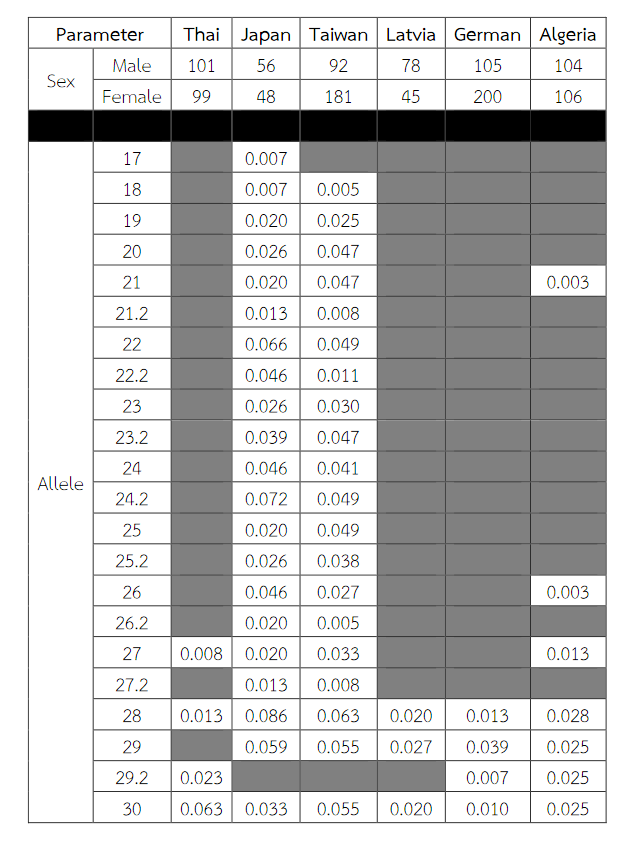
Table 9 (Continued) Allele frequencies, statistical parameters, and references of DXS10011 in six populations. For the Thai population, the data were from pooled males and females (Table 3) when the exact tests showed no difference between the two groups. No significant most common allele shared in any population was observed. Some statistical parameters were not available.
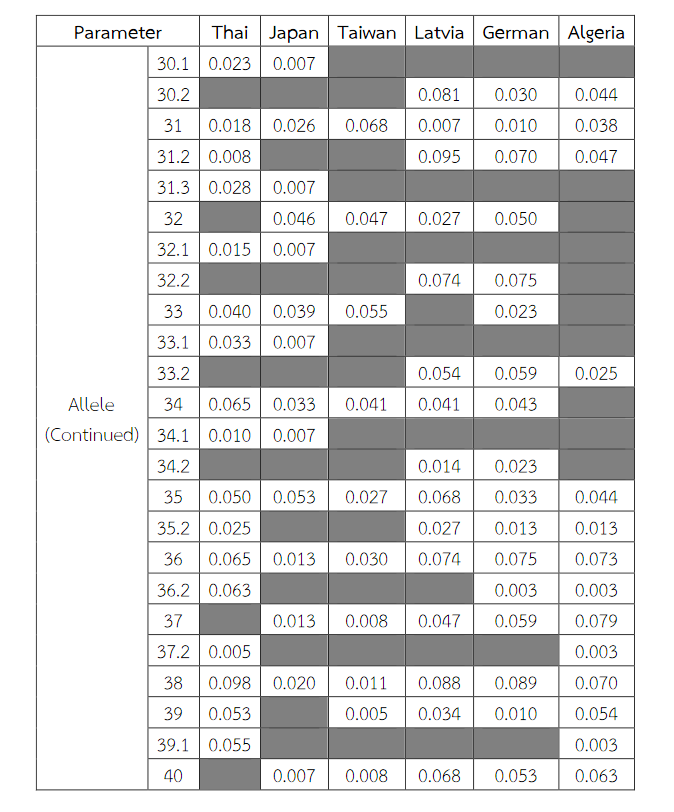
Table 9 (Continued) Allele frequencies, statistical parameters, and references of DXS10011 in six populations. For the Thai population, the data were from pooled males and females (Table 3) when the exact tests showed no difference between the two groups. No significant most common allele shared in any population was observed. Some statistical parameters were not available.
Discussions
Individuals from the Thai population including 101 males, 99 females and 15 family trios with female children were successfully investigated with five X chromosome markers. The multiplex PCR assays were optimized for a DNA concentration of 4 ng initially but later were able to be diluted to 2 ng, that is, they were sensitive enough for routine paternity analysis. The DXS10011 locus was highly polymorphic, with the highest power of discrimination and probability of paternity exclusion among the five markers studied.
When the comparison of allele frequencies between different races was observed, the Asian population shared the most common allele, that is, allele 10 at locus DXS8378 and allele 13 at locus HPRTB whereas allele 49 at locus DXS8377 was a most common allele shared among the African population. DXS10011 had the greatest individual value as a forensic marker, with the highest power of discrimination in males (PDm) and females (PDf), as well as mean exclusion chance.
DXS10011 had a very low HETobs (0.670) compared to the expected heterozygosity (HETexp) (0.950) as a result of an amplification difficulty, leading to a genotypic error (homozygote excess)(12). It also had the largest product size compared to DXS8378, DXS101, HPRTB, and DXS8377(4,13-15). In addition, DXS10011 contained a highly polymorphic allele, mainly because of complex structural variants (regular and inter-alleles)(14). We found that the shorter fragment size amplified better than the larger fragment which produced allelic drop-out, known as extreme preferential amplification (EPA). Due to a difficulty to distinguish the longer allele from background noise, the overrepresentation of homozygotes for the shorter allele could occur(12). Therefore, some samples were repeatedly investigated.
Because the recombination rate of the X chromosome in females is low, the X chromosome has greater linkage disequilibrium (LD) as compared to autosomal markers. The exact test for LD was performed for all pairs of the loci in this study, the result showed no LD in each pair of markers. In the kinship cases involving 15 family trios with daughter, no mutation was detected. This proved the applicable of five X-STR markers in kinship cases.
Conclusions
This five X-chromosomal STRs multiplex system offered sufficient polymorphic patterns in one single reaction. It worked with reasonable amounts of DNA, suitable for forensic casework, and yielded reproducible results. It could be recommended for routine paternity analysis in complex deficiency cases or for a complement of autosomal and Y-STRs analysis.
Acknowledgements
The authors would like to thank Ms Janpen Thanakitgosate, Ms Ubonrat Jomsawat and Ms Jittima Shotivaranon for their generous technical support on DNA amplification and DNA fragment analysis. This work was supported by a grant from the Faculty of Medicine Ramathibodi Hospital, Mahidol University.
References
- B. Brinkmann, Overview of PCR-based systems in identity testing, Methods Mol. Biol. 198 (1998) 105–119.
- M. Kayser, A. Caglia´, D. Corach, N. Fretwell, C. Gehrig, G. Graziosi, F. Heidorn, S. Herrmann, B. Herzog, M. Hidding, K. Honda, M. Jobling, M. Krawczak, K. Leim, S. Meuser, E. Meyer, W. Oesterreich, A. Pandya,W. Parson, G. Penacino, A. Perez-Lezaun, A. Piccinini, M. Prinz, C. Schmitt, P.M. Schneider, R. Szibor, J. Teifel-Greding, G.Weichhold, P. de Knijff, L. Roewer, Evaluation of Y-chromosomal STR's: a multicenter study, Int.J.Legal Med. 110 (1997) 125–133.
- J. Edelmann, R. Szibor, Validation of the HumDXS6807 short tandem repeat polymorphism for forensic application, Electrophoresis 20 (1999) 2844–2846.
- J. Edelmann, R. Szibor, DXS101: a highly polymorphic X-linked STR, Int. J. Legal Med. 114 (2001) 301–304.
- R. Szibor, M. Krawczak, S. Hering, J. Edelmann, E. Kuhlisch,D. Krause, Use of X-linked markers for forensic purposes, Int.J.Legal Med. 117 (2003) 67–74.
- S. Turrina, D. De Leo, Population data of three X-chromosomal STRs: DXS7132, DXS7133 and GATA172D05 in North Italy, J. Forensic Sci. 48 (2003) 1428–1429.
- M. Lv, W. Liang, M. Wu, M. Liao, B. Zhou, Y. Jia, L. Zhang, Allele frequency distribution of two X-chromosomal STR loci in Han population in China, J. Forensic Sci. 49 (2004) 418–419.
- M. Ross, D. Grafham, A. Coffey, The DNA sequence of the human X chromosome, Nature 434 (2005) 325–337.
- R.H. Don, P.T. Cox, B.J. Wainwright, K. Baker, J.S. Mattick, ''Touch-down'' PCR to circumvent spurious priming during gene amplification, Nucleic Acids Res. 19 (1991) 4008.
- DNA Commission of the International Society of Forensic Hemogenetics, DNA recommendations-further report of the DNA commission of the ISFH regarding the use of short tandem repeat systems, Int.J.Legal Med 110 (1997) 175–176.
- R. Fisher, Standard calculations for evaluating a blood group system, Heredity 5 (1951) 95–102.
- J.J. Chen, T. Duan, R. Single, K. Mather, G. Thomson, Hardy-Weinberg testing of a single homozygous genotype, Genetics 170 (2005) 1439-1442.
- S. Hering, N. Brundirs, E. Kuhlisch, J.Edelmann, I. Plate, M. Benecke, P.H. Van, M. Micheal, R.Szibor, DXS10011: studies on structure, allele distribution in three populations and genetic linkage to further q-telomeric chromosome X markers, Int.J.Legal Med 118 (2004) 313-319.
- J. Edelmann, D. Deichsel, S. Hering, I. Plate, R. Szibor, Sequence variation and allele nomenclature for the X-linked STRs DXS9895, DXS8378, DXS7132, DXS6800, DXS7133, GATA172D05, DXS7423 and DXS8377, Forensic Sci.Int.129 (2002) 99-103.
- M. Poetsch, H. Petersmann, A. Repenning, E. Lignitz, Development of two pentaplex systems with X-chromosomal STR loci and their allele frequencies in a northeast German population, Forensic Sci.Int. 155 (2005) 71-76.
- D. Athanasiadou, B. Stradmann-Bellinghausen, C. Rittner, K.W. Alt, P.M. Schneider, Development of a quadruplex PCR system for the genetic analysis of X-chromosomal loci, Int. Congress Series 1239 (2003) 311-314.
- H. Asamura, H. Sakai, M. Ota, M. Fukushima, Japanese population data for eight X-STR loci using two new quadruplex systems, Int J Legal Med. 120 (2006) 303–309.
- H. Asamura,H. Sakai,K. Kobayashi,M. Ota, M. Fukushima, MiniX-STR multiplex system population study in Japan and application to degraded DNA analysis, Int.J.Legal Med. 120 (2006) 174–181.
- A. Tamura, M. Iwata, I. Takase, T. Miyazaki, K. Matsui, H. Nishio, K. Suzuki, Analysis of two types of novel alleles in the DXS10011 locus, Legal Medicine. 6 (2004) 52–54.
- W.M. Tang, K.Y.To, Four X-chromosomal STRs and their allele frequencies in a Chinese population, Forensic Sci.Int. 162 (2006) 64–65.
- M. Chen, C. Pu, Population data on the X chromosome short tandem repeat loci DXS10011, DXS101, DXS6789, DXS7132, DXS8377and DXS9895 in Taiwan, Forensic Sci.Int. 146 (2004) 65–67.
- S. Turrina, R. Atzei, G. Filippini, D. De Leo, Development and forensic validation of a new multiplex PCR assay with 12 X-chromosomal short tandem repeats, Forensic Sci.Int.: Genetics. 1 (2007) 201–204.
- C. Robino, A. Giolitti, S. Gino, C. Torre, Analysis of 12 X-chromosomal short tandem repeats in the North-West Italian population by means of two multiplex PCRs, Int. Congress Series. 1288 (2006) 316–318.
- M. Poetsch, A. Sabule, H. Petersmann, V. Volksone, E. Lignitz, Population data of 10 X-chromosomal loci in Latvia, Forensic Sci.Int. 157 (2006) 206–209.
- I. Gomes, C. Alves, K. Maxzud, R. Pereira, M.J. Prata, P. Sa´nchez-Diz, A. Carracedo, A. Amorim, L. Gusma˜o, Analysis of 10 X-STRs in three African populations, Forensic Sci.Int.: Genetics. 1 (2007) 208–211.
- A. Bekada, S. Benhamamouch, A. Boudjema, M. Fodil, S. Menegon, C. Torre, C. Robino, Analysis of 12 X-chromosomal STRs in an Algerian population sample, Forensic Sci.Int.: Genetics Supplement Series. 2 (2009) 400–401.


